Fruiting Body Dna Rna Extraction
title: Extracting DNA and RNA from Fruiting Bodies tags:
- dna extraction
- rna extraction
- fruiting body author: Annegret Kohler, Claude Murat, Francis Martin member: annegret-kohler —
EXTRACTING DNA AND RNA FROM FRUITING BODIES
by Annegret Kohler, Claude Murat & Francis Martin at INRA-Nancy
Getting started
A review about JGI sample preparation requirements can be found here. By October 2014 a fungal standard draft project requires 12µg of high quality DNA and 3µg of total RNA. So ideally you should harvest a big enough specimen of excellent quality or if this is not possible several specimens from the same site.
Plan your field visit
Document your samples by taking photographs of the sampled fruiting bodies. A detailed written description of each samples is a must. Record the GPS coordinates of the site.
If you take the time to seek out mushrooms for genome sequencing, it only makes sense to exercise the extra care and trouble necessary to get them to the lab in excellent conditions. Always use sample young specimens and fresh material. Specimens should be frozen as quickly as possible on site. We recommend to use dry ice, which is relatively easy to transport and to handle in the forest (of course by respecting the necessary safety arrangements). Other possibilities would be liquid nitrogen or RNAlater (not working for all kind of samples, it has to be tested in advance). In addition to dry ice you will need tools to cut and store the fruiting bodies (scalpel, gloves, suitable tubes….). Cut the material already in small pieces before freezing, this will facilitate the homogenization of the sample prior to extraction. Back to the lab, transfer the samples rapidly to -80°C! If immediate freezing of the materiel is not possible, transport the fruiting bodies to the lab and freeze them asap.
Use only clean parts of the fruiting body (e.g., central part of the upper stipe) and avoid parts with obvious contamination (insect larvea, slugs, etc.) and gills containing spores. Keep a tissue sample for rDNA ITS identification. It is highly recommended to keep part of the fruiting body for a voucher and/or keep unfrozen material to try culturing on agar plates back to the lab.
RNA extraction
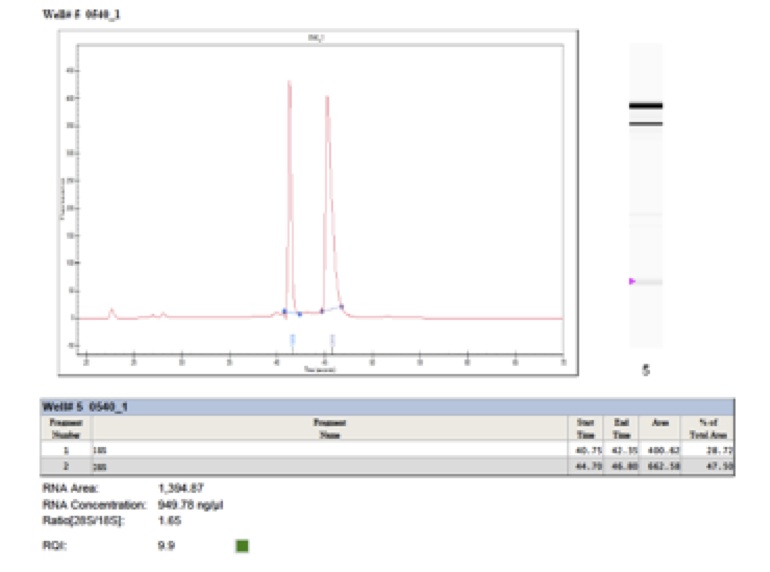
Commercial kits usually work well for fruiting bodies. We recommend RNeasy Plant Mini Kit (Qiagen) by following the protocol for filamentous fungi and using RLC buffer. Don’t forget to include a DNase treatment. Use Bioanalyzer and Qubit (or similar equipment) to quantify your RNA and to control the quality.
DNA extraction of genomic DNA
We have mainly used two protocols to extract high quality genomic DNA from fresh fruiting bodies. The first protocol is a slightly modified PowerMax Soil DNA Isolation Kit (MoBio 12988-10) PowerMax Protocol PDF; the second protocol is based on the CTAB extraction protocol DNAProtocol AK0511 PDF, combined with genomic tip columns (Qiagen) for purification.
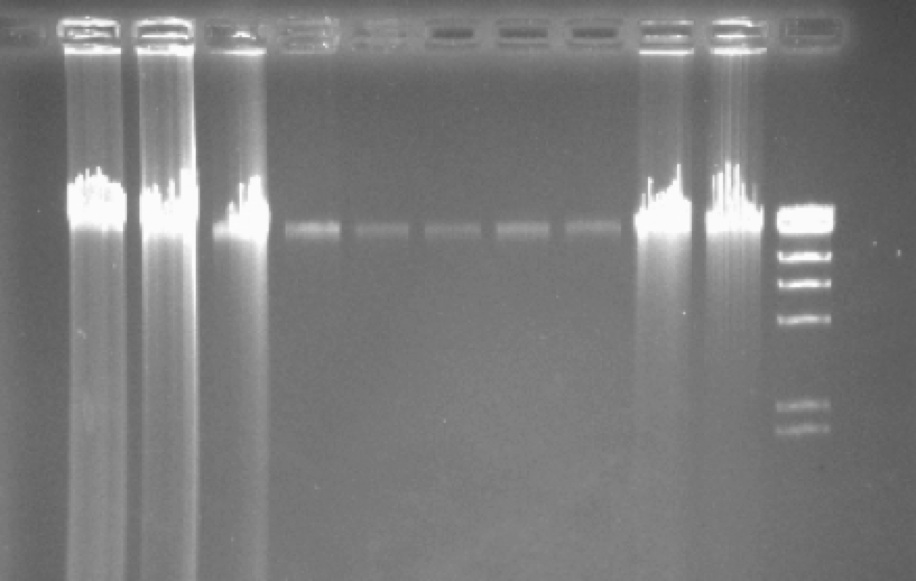
After DNA extraction prepare a 1% agarose gel and load 1-5µl of the genomic DNA extract (eventually several dilutions), together with a DNA size marker to control the quality of your DNA. Use Qubit (Invitrogen) to quantify your sample. Avoid the NanoDrop assay.
Confirm the taxonomic attribution
Before sending the DNA for sequencing, confirm the species attribution by sequencing the rDNA ITS and querying the UNITE database.
Assessing bacterial contamination
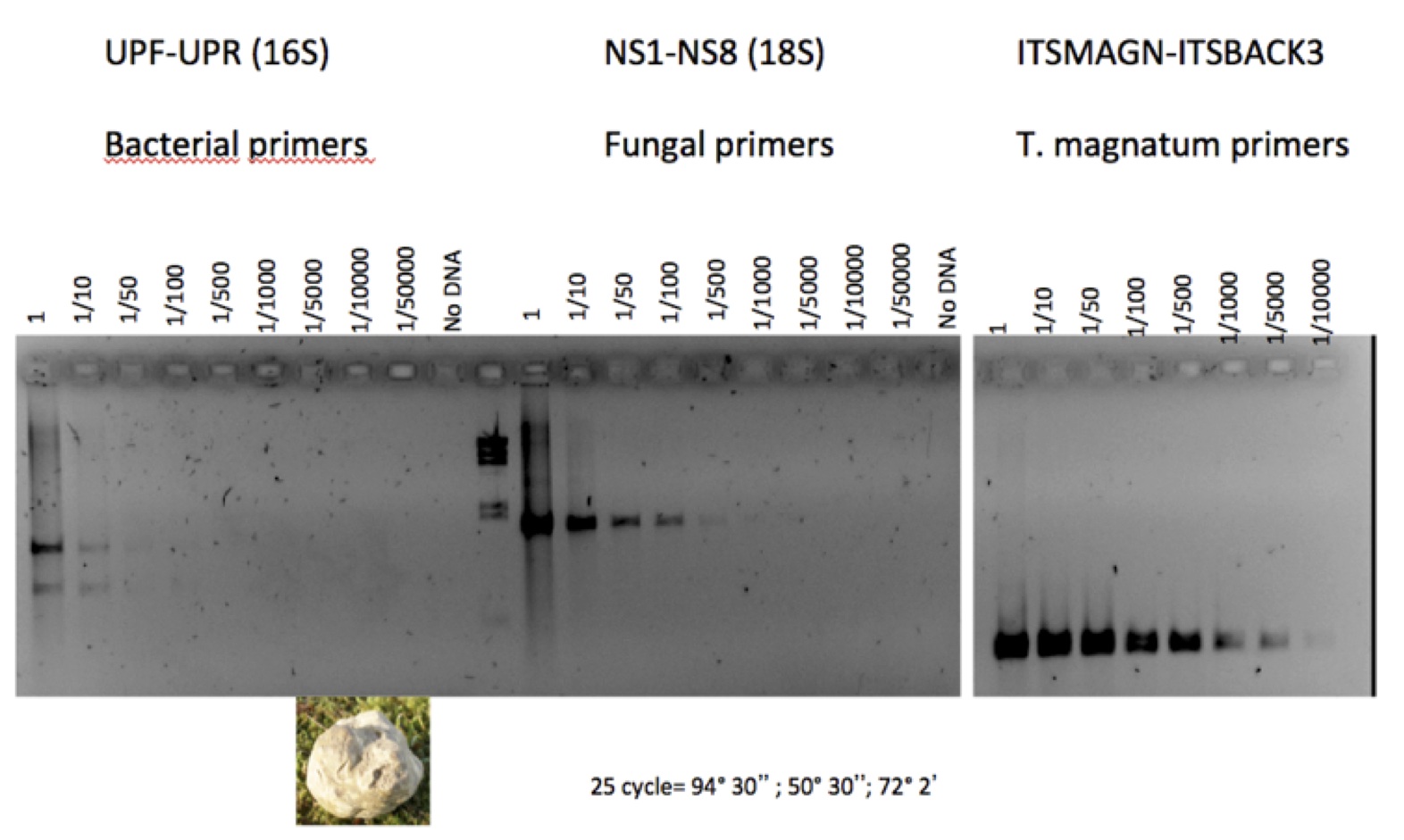
Fruiting body DNA are frequently contaminated by large amount of bacterial DNA. To assess the degree of contamination a semi-quantitative PCR test is applied by using 16S and 18S primer (to amplify bacterial and fungal DNA), see below for an assay carried out on Tuber magnatum gDNA). DNA samples with >15-20% bacterial DNA should be discarded.
Ship genomic DNA to the sequencing center following their recommended procedures, but always keep a DNA aliquot at -80°C for further analysis (if needed).
PCR primer sequences used for assessing bacterial contamination of fungal gDNA:
Bacterial 16S rDNA:
UPForward AGAGTTTGATYMTGGC
UPReverse GYTACCTTGTTACGACTT
Fungal 18S rDNA :
NS1 GTAGTCATATGCTTGTCTC
NS8 TCCGCAGGTTCACCTACGGA
Fungal rDNA ITS :
ITS1F CTTGGTCATTTAGAGGAAGTAA
ITS4 TCCTCCGCTTATTGATATGC
Previous post
Graduate student-postdoc challenge
Next post
1000 Genomes Progress (2018 update)